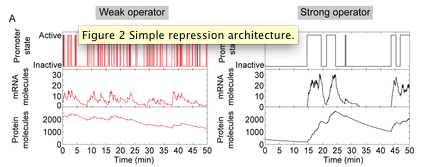
In a recently published paper “Effect of promoter architecture on the cell-to-cell variability in gene expression” in PLoS Computational Biology, Alvaro Sanchez and co-workers investigated how the architecture of a model promoter region (characterized by number of transcription factor binding sites, the binding affinity and spacing on the DNA) affects the way in which individual cells respond to environmental stimuli. In particular, they examine, using stochastic chemical kinetics, how the intrinsic randomness in the binding and unbinding of transcription factors to their binding sites generates cell-to-cell differences in transcript and protein levels within a population. The analysis uses a combination of computational modeling and analytical mathematical methods. Sanchez, a recent Ph.D. graduate in Biophysics and Structural Biology performed this work with Jané Kondev (Physics), and in collaboration with Rob Phillips, Hernan Garcia and Daniel Jones (Caltech).
While previous population-average models explained well how promoter architecture affects the average response of a population of cells to changes in the concentration of transcription factors, the question of how the response of individual cells is determined by promoter region sequence remains generally unsolved and limited to simplified coarse-grain models. By way of an example, the authors of this study investigated the effect of cooperative binding between transcription factors in the level of variability in the transcriptional response to increasing concentrations of those factors. It is well known that cooperativity in gene regulation increases the sensitivity of the response of the promoter to changes in the intracellular concentration of transcription factors, leading to a switch-like response. By examining this architecture, Sanchez and co-workers found that cooperativity is also a source of large intrinsic cell-to-cell variability in gene expression: larger sensitivity comes accompanied with larger variability (even if all cells contain the exact same amount of repressor).
This investigation continues a collaboration between theorists at Brandeis and experimentalists at Caltech, which aims to connect the biochemical, molecular understanding of transcriptional regulation coming from in vitro biochemical experiments (which are also being done in the Gelles lab at Brandeis) with the phenotypic behavior of individual cells as determined by gene expression measurements in single live cells. Many of the predictions of this computational study are currently being tested in Rob Phillips’ lab.