 by Zachary Knecht, PhD candidate
by Zachary Knecht, PhD candidate
As the solvent of living cells, water is critical for all life on earth. This makes monitoring how environmental conditions impact evaporation and subsequently sensing and locating water sources important for animal survival. This is particularly critical for insects, whose small body size makes them highly susceptible to dehydration. In addition, moisture sensing, or hygrosensation, is also important for the spread of insect-born disease. Mosquitoes that spread malaria or viruses like dengue and Zika, not only need to locate bodies of standing water in which to lay eggs, but also home in on the moisture that emanates from our bodies when searching for a blood meal. This dual role for hygrosensing in mosquito biology makes their hygrosensory machinery a promising target for pest control strategies. Until now though, the genes and molecules that function in insect hygrosensation have been completely unknown.
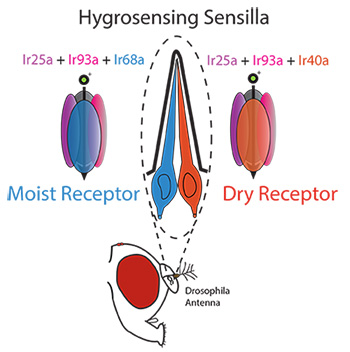
In a pair of recent papers in the journal eLife, researchers in the Garrity Lab at Brandeis University, in collaboration with colleagues at the University of Lausanne in Switzerland, have uncovered the cellular and molecular mechanisms that underlie insect hygrosensation using the fruit fly Drosophila melanogaster. Like mosquitoes, fruit flies detect humidity through specialized, innervated hair-like structures located on their antennae called sensilla. Each hygrosensing sensilla contains one cell that responds to increasing humidity (a moist cell), and one that responds to decreasing humidity (a dry cell). These papers demonstrate that the balance of activity between dry and moist cells allows the insect to seek out or avoid particular humidity levels, a preference which changes depending on how hydrated or dehydrated the fly is.
To identify the molecules involved in sensing moisture, the researchers looked for mutant flies unable to distinguish between humid and dry air. They found that animals with mutations in four different genes disrupted the behavior. Strikingly, each of these genes encoded a different member of the same family of sensory receptors, the so-called Ionotropic Receptors or IRs. Although IRs are found only in invertebrates, they belong to the same family as the ionotropic Glutamate Receptors, which lie at the heart of communication between nerve cells in the animal brain, including the human brain. IRs differ from these relatives in that instead of sensing signals sent by neurons, they detect signals coming from the environment. IRs are best known to act as chemical receptors, but the group found that a subset of IRs act instead to sense humidity. The researchers found two broadly expressed IRs, Ir25a and Ir93a, were required by both the dry cells and moist cells while the other two IRs, Ir40a and Ir68a, were specifically required by the dry and the moist cells, respectively. This suggests that Ir25a and Ir93a contribute to the formation of both moist and dry receptors, while Ir40a and Ir68a provide the dry- and moist-specific subunits to the receptor. Consistent with this view, the loss of either Ir68a or Ir40a alone only partially reduces the animal’s ability to sense humidity, but animals with mutations in Ir25a, Ir93a or both Ir40a and Ir68a are completely blind to moisture.
Having identified the specific genes required for sensing moisture, the next step is to determine the precise mechanism by which humidity activates these receptors. Furthermore, these genes are conserved in mosquitoes and other disease vectors, providing a clear path to translate what’s known about fly hygrosensation into the mosquito. These papers lay the groundwork for new mosquito control strategies that aim to precisely inhibit their ability to seek out water to reproduce and to seek out hosts to bite and spread deadly pathogens.





